About MYCOsiRNA
Limitations of past methods:
Gene silencing through RNA interference (RNAi) is a widely used molecular tool for functional genomics. The biogenesis of siRNA and its binding to the target for gene silencing is a multi-step process of RNA interference (RNAi) pathways. Although a number of siRNA design tools have been developed for animals and plants, there is still no available tool for designing effective, specific, and non-toxic fungi and oomycete siRNAs against a target gene.
Novelty:
We present
MYCOsiRNA, a web server tool to design effective, specific and non-toxic siRNAs for fungi and oomycete RNAi.
Screening criteria for highly effective and specific
fungal and oomycetous siRNA:
- Candidate siRNAs containing four consecutive A, U, C or G will be discarded.
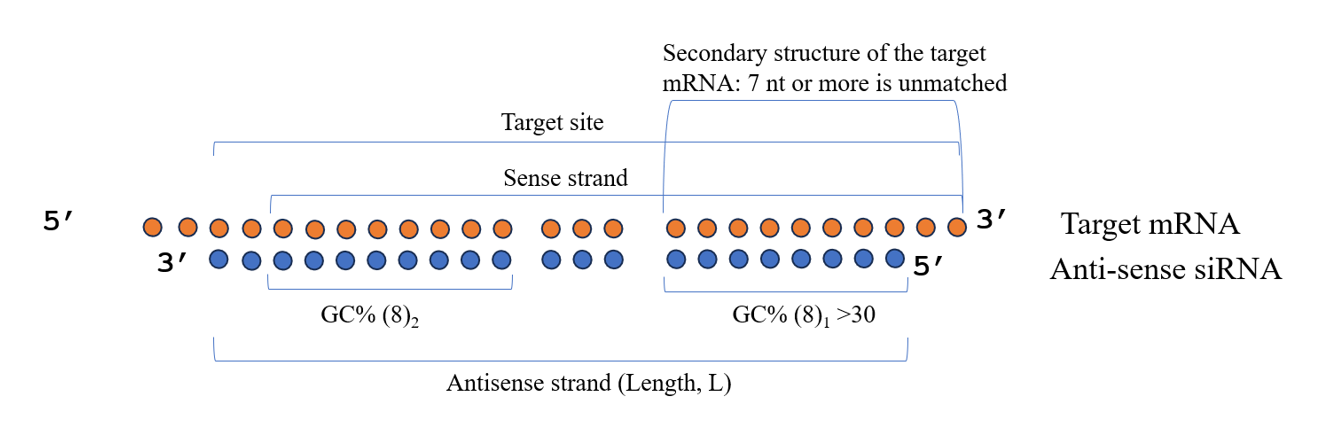
- GC content of the antisense strand is between 40% and 60%.
- GC% (8)1 > 30% and GC% (8)2 > GC% (8)1
- Target accessibility: 70% or more of the last 10 nucleotides of the target site are unmatched, indicated by “.”.
- Off-target genes are defined as genes to which 13 or more consecutive nucleotides of the siRNA could bind. Candidate siRNAs with more than five off-target genes will be discarded.